Overview of SARS-CoV-2 Viral Entry
The generally accepted model for SARS-CoV-2 viral entry involves two crucial elements: the Spike protein on the surface of the enveloped virus, and the host receptor ACE2. The Spike protein has a high affinity for the host ACE2, and it binds directly to it, leading ultimately to internalization of the virus. The internalization appears to be triggered by proteases that cut the Spike protein in two places. The SARS-CoV-2 virus that causes Covid-19 has a new multibasic site adjacent to one of the cleavage sites, and there is now evidence that the multibasic site can interact with a different host receptor, neuropilin 1 (Daly et al. 2020). Three different proteases appear to be involved in the processing and activation of Spike. One is furin, thought to be involved in the processing of the Spike protein during viral replication (Shang et al. 2020). Another is TMPRSS2, a host protease on the cell surface (Matsuyama et al. 2020; Shirato, Kawase, and Matsuyama 2018), and finally Cathepsin L, a pH dependent lysosomal protease (Huang et al. 2006; Ou et al. 2020). These mechanisms of viral entry are the basis of our pseudovirus assays.
Detecting Viral Entry in Live “Host” Cells
Our fluorescent viral entry assay has two components. The first is a red fluorescent protein fused to the C-terminus of Human ACE-2, packaged in a BacMam vector. The BacMam vector is pseudotyped with VSVG for efficient delivery to mammalian “host” cells. For example, A549 cells expressing red fluorescent ACE-2, become permissive to entry by SARS-CoV-2 or pseudo-SARS-CoV-2. To quantify ACE-2 mediated entry, adjacent wells of cells are transduced with either red fluorescent ACE2 or a red fluorescent control. The control is a BacMam vector that only expresses a red cytosolic fluorescent protein. The red fluorescent ACE-2 reporter and control are available in the same kit, product number #C1100R.
The second assay component is a “pseudovirus” that enables research in BSL1/BSL2 facilities #C1110G. This pseudovirus is a modified BacMam that presents SARS-CoV-2 Spike protein on the surface of the BacMam capsid. The Spike protein is derived from the original Wuhan isolate. Wild type Spike protein has an ER retention motif (McBride, Li, and Machamer 2007), that we removed to improve presentation on the pseudovirus surface. We appended 7 amino acids from the GP64 glycoprotein protein (Yang et al. 2007). Following host cell entry, the pseudovirus expresses bright green fluorescence in the “host” cell nucleus. A bald pseudovirus control has neither Spike nor VSVG on the surface, but expresses nuclear targeted mNeonGreen just as the pseudo virus does. This control indicates basal infectivity from BacMam glycoprotein GP64. ACE-2/Spike mediated entry can be quantified relative to the control.
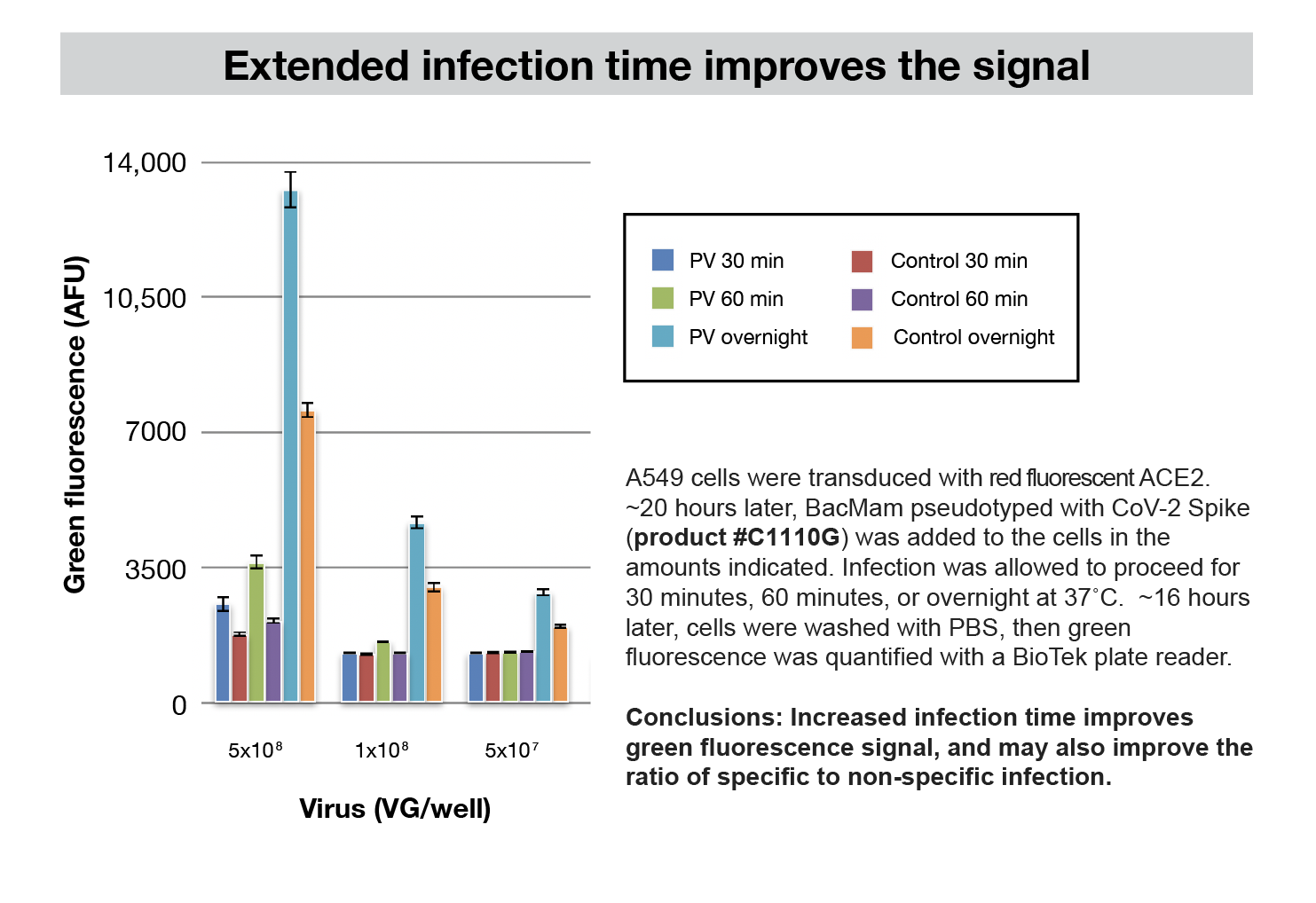
Other Cell Types
This assay works well an A549 cell line with both SARS-CoV-2 and the BacMam pseudovirus. A549 cells are a human epithelial lung cell line. It is conceivable that these assays will be useful in other cell types, but there are several host proteins involved in the SARS-CoV-2 viral entry, and the assay may need further optimization in other cell types. Early adopters are now testing Vero cells and other cell types. We’ll provide further information about new developments. We’re here to help, and appreciate hearing from the community.
Montana Molecular COVID-19 Reagent Pipeline
A new pseudovirus carrying the D614G Spike which originally appeared in a European isolate is in production. This mutation appears to have functional consequences for viral entry (Korber et al. 2020; Zhang et al. 2020). QA/QC for the D614G mutant is scheduled for mid July. A green fluorescent Mpro protease sensor is scheduled to ship in early July.
Daly, James L., Boris Simonetti, Carlos Anton Plagaro, Maia Kavanagh Williamson, Deborah K. Shoemark, Lorena Simon-Gracia, Katja Klein, et al. 2020. “Neuropilin-1 Is a Host Factor for SARS-CoV-2 Infection.” bioRxiv. https://doi.org/10.1101/2020.06.05.134114.
Huang, I-Chueh, Berend Jan Bosch, Fang Li, Wenhui Li, Kyoung Hoa Lee, Sorina Ghiran, Natalya Vasilieva, et al. 2006. “SARS Coronavirus, but Not Human Coronavirus NL63, Utilizes Cathepsin L to Infect ACE2-Expressing Cells.” The Journal of Biological Chemistry 281 (6): 3198–3203.
Korber, B., W. M. Fischer, S. Gnanakaran, H. Yoon, J. Theiler, W. Abfalterer, B. Foley, et al. 2020. “Spike Mutation Pipeline Reveals the Emergence of a More Transmissible Form of SARS-CoV-2.” bioRxiv. https://doi.org/10.1101/2020.04.29.069054.
Matsuyama, Shutoku, Naganori Nao, Kazuya Shirato, Miyuki Kawase, Shinji Saito, Ikuyo Takayama, Noriyo Nagata, et al. 2020. “Enhanced Isolation of SARS-CoV-2 by TMPRSS2-Expressing Cells.” Proceedings of the National Academy of Sciences of the United States of America 117 (13): 7001–3.
McBride, Corrin E., Jie Li, and Carolyn E. Machamer. 2007. “The Cytoplasmic Tail of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Contains a Novel Endoplasmic Reticulum Retrieval Signal That Binds COPI and Promotes Interaction with Membrane Protein.” Journal of Virology 81 (5): 2418–28.
Ou, Xiuyuan, Yan Liu, Xiaobo Lei, Pei Li, Dan Mi, Lili Ren, Li Guo, et al. 2020. “Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and Its Immune Cross-Reactivity with SARS-CoV.” Nature Communications 11 (1): 1620.
Shang, Jian, Yushun Wan, Chuming Luo, Gang Ye, Qibin Geng, Ashley Auerbach, and Fang Li. 2020. “Cell Entry Mechanisms of SARS-CoV-2.” Proceedings of the National Academy of Sciences of the United States of America, May. https://doi.org/10.1073/pnas.2003138117
Shirato, Kazuya, Miyuki Kawase, and Shutoku Matsuyama. 2018. “Wild-Type Human Coronaviruses Prefer Cell-Surface TMPRSS2 to Endosomal Cathepsins for Cell Entry.” Virology 517 (April): 9–15.
Yang, Ding-Gang, Yao-Chi Chung, Yiu-Kay Lai, Chia-Wei Lai, Hung-Jen Liu, and Yu-Chen Hu. 2007. “Avian Influenza Virus Hemagglutinin Display on Baculovirus Envelope: Cytoplasmic Domain Affects Virus Properties and Vaccine Potential.” Molecular Therapy : The Journal of the American Society of Gene Therapy 15 (5): 989–96.
Zhang, Lizhou, Cody B. Jackson, Huihui Mou, Amrita Ojha, Erumbi S. Rangarajan, Tina Izard, Michael Farzan, and Hyeryun Choe. 2020. “The D614G Mutation in the SARS-CoV-2 Spike Protein Reduces S1 Shedding and Increases Infectivity.” bioRxiv. https://doi.org/10.1101/2020.06.12.148726.
Huang, I-Chueh, Berend Jan Bosch, Fang Li, Wenhui Li, Kyoung Hoa Lee, Sorina Ghiran, Natalya Vasilieva, et al. 2006. “SARS Coronavirus, but Not Human Coronavirus NL63, Utilizes Cathepsin L to Infect ACE2-Expressing Cells.” The Journal of Biological Chemistry 281 (6): 3198–3203.
Menachery, Vineet D., Kenneth H. Dinnon 3rd, Boyd L. Yount Jr, Eileen T. McAnarney, Lisa E. Gralinski, Andrew Hale, Rachel L. Graham, et al. 2020. “Trypsin Treatment Unlocks Barrier for Zoonotic Bat Coronavirus Infection.” Journal of Virology 94 (5). https://doi.org/10.1128/JVI.01774-19.
Ou, Xiuyuan, Yan Liu, Xiaobo Lei, Pei Li, Dan Mi, Lili Ren, Li Guo, et al. 2020. “Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and Its Immune Cross-Reactivity with SARS-CoV.” Nature Communications 11 (1): 1620.
"entry" - Google News
June 27, 2020 at 06:15AM
https://ift.tt/3dA7bbX
SARS-CoV-2 Pseudovirus Entry Assay - Montana Molecular
"entry" - Google News
https://ift.tt/3f5ZAUJ
https://ift.tt/3d6LMHD
Bagikan Berita Ini














Excellent post!
ReplyDelete